 CAP Update December 2012: RNAi for Treating Honey Bee Diseases
CAP Update December 2012: RNAi for Treating Honey Bee Diseases
RNAi for Treating Honey Bee Diseases
Author: Yanping (Judy) Chen and Jay D. Evans, USDA-ARS Beltsville Bee Research Laboratory, Beltsville, MD 20705
Originally Jointly Published: American Bee Journal and Bee Culture, Decemeber 2012
Background
Many ground-breaking discoveries in science have occurred through serendipity. RNA interference (RNAi), a natural process to turn off gene activity in plants and animals is one of the many serendipitous discoveries that involved a mix of unexpectedness and insight that led to a valuable outcome. Back in the late 1980s, Dr. Rich Jorgenson, a molecular geneticist, and his colleague were trying to create a petunia with intensely purple colored flowers by inserting multiple copies of a purple color-producing gene into petunia plants. Surprisingly, the result turned out to be completely different from their expectation. Instead of achieving darker colored flowers, they got plants with white or patchy blossoms. At the time, no one quite understood why the addition of extra copies of an introduced gene silenced both themselves and the plant’s own purple color producing genes. Later, Drs. Andrew Fire and Craig Mello pieced together the molecular machinery behind color alternations in petunias and identified an efficient mechanism of cells to regulate protein production at the RNA stage, a mechanism famously known as RNAi (Fire et al., 1998). The discovery of RNAi has been viewed as a major breakthrough in cell biology and Drs. Fire and Mello were awarded the Nobel Prize in Physiology or Medicine 2006 for their discovery of RNAi.
RNAi Mechanism and Application
A gene is the functional unit of heredity in all living organisms. It consists of a specific section of DNA that holds the genetic information that each organism uses to pass genetic traits onto their offspring. The information held in this gene is converted into a protein that the organism can use in two steps. First, messenger RNA (nearly a mirror image of the gene) is produced after cues received by the organism’s cells. This RNA is then used to direct the production of new proteins, the long stretches of amino acids that build bodies and actions.
RNAi is one tool used by cells to turn up or down the production of specific proteins. It is used both to weak protein levels of the host itself and to knock down proteins from viruses and other agents that attack the host. As the Nobel prize winners discovered, the RNAi pathway is triggered by the presence of double-stranded RNA (dsRNA), an unusual event whereby RNA adheres to an exact mirror of itself. These dsRNA strands are recognized as being odd, and are consequently recognized by the appropriately named protein ‘Dicer’. Dicer binds and cleaves long stretches of dsRNA into short fragments which are called short interfering RNA (siRNA).
Diced siRNA products are carried through the cell by a protein complex, finding their own matches in the RNA soup and degrading these matches. This degraded mRNA can no longer serve as a template for protein synthesis and therefore a specific gene’s activity is silenced (Figure 1).
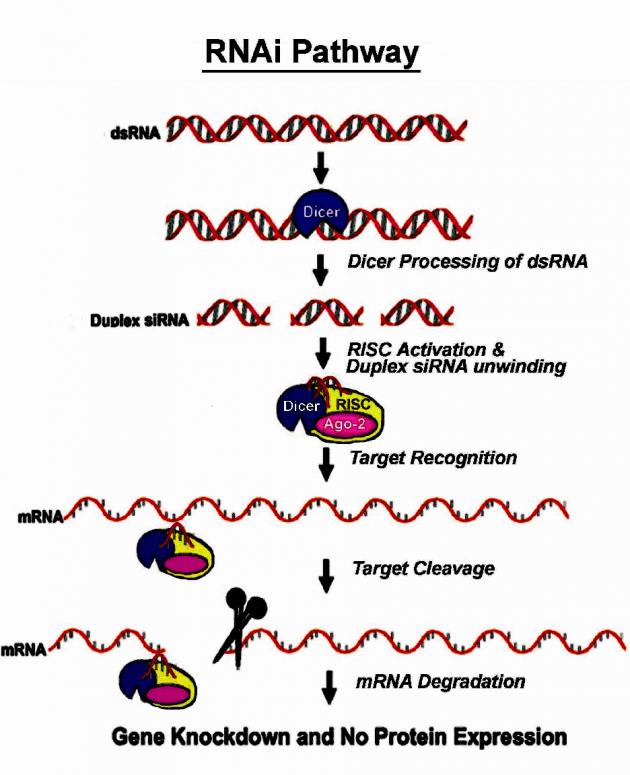
Figure 1: Graphical representation of RNAi pathway
The ability to target and switch off specific genes with high sequence specificity makes RNAi a powerful tool for studying gene function and a promising approach for treating a variety of diseases. Since RNAi can specifically inhibit the function of any chosen target genes, this technique can theoretically treat diseases that are caused by errant gene expression and protein function. In addition, it is known that RNAi has an appetite for RNA viruses along with other pathogens and parasites. In fact, the RNAi system might be maintained in large part as a defense against the many RNA viruses found in plants and animals. In order to reproduce, these viruses pass through a dsRNA stage, and that is when they appear on the Dicer’s radar.
RNAi Application in Honey Bee Disease Treatment
Honey bees possess the core components of the RNAi pathway including Dicer and the enzymes and structural proteins that use ‘diced’ RNAs to hunt down and degrade matching RNA (review in Aronstein et al. 2011). As a result, RNAi looks promising as a tool for combating honey bee pathogens and parasites. Beeologics, a company focused on the discovery, development, and commercialization of RNAi therapeutics (recently acquired by Monsanto), has developed a product called Remebee, a dsRNA homologous to Israeli acute paralysis virus (IAPV) that was found to be associated with honey bee Colony Collapse Disorder (CCD) (Cox-Foster et al. 2007). The injection and feeding of Remembee has proven effective in reducing the abundance of IAPV in bees and promoting bee health under both laboratory (Maori et al., 2009) and large-scale natural beekeeping conditions (Hunter et al., 2010). Additionally, Beeologics scientists, along with USDA collaborators, have shown that RNAi can be directed at reducing the gut parasite, Nosema ceranae (Paldi et al., 2010).
Recently, with CAP support, we conducted studies to further elucidate the antiviral effect of RNAi on the infection and replication of IAPV in honey bees. We targeted the IRES region of this virus, a key stretch needed to exploit the bees’ own protein-making factory, the ribosome. To generate what we hoped was a more potent trigger for RNAi, we jumped past the dicing step by providing bees with siRNA that was ready to work as a bait for targeting viruses. In our trials, the frames with emerging brood were removed from the colonies identified with IAPV infection by RT-PCR assay and newly emerged bees were collected at the following day. Thirty newly emerged bees were placed into multiple rearing cages (Evans et al. 2009) as part of four groups. A scintillation vial filled with a1:1 sugar syrup: water mix (10 ml) was inverted over the top of the rearing cup to provision caged bees. Varroa mites were collected by sugar roll from a colony that was identified to have IAPV infection. Two groups were challenged with Varroa mites from an IAPV-rich colony, with one of these groups also receiving an siRNA cocktail directed at the IAPV IRES region (Treatment 1) and one receiving no siRNA (Negative control 1). Two groups were kept mite-free, with one of these receiving siRNA (Treatment 2) and one not (Negative Control-2). Each group consisted of four replicates. After setting up the experiment at day 1, the bees were collected at three-day intervals (day 4, day 7, and day 10) individually. We then measured virus levels using standard genetic techniques (Figure 2). The trial was repeated three times.

Figure 2: Bees contained in vials, separated by treatment and replicate
The results showed that the feeding of siRNA targeting IAPV could confer antiviral activity in bees (Figure 3). The IAPV infection rates in bees from Treatment-I and Treatment-II, which were fed with siRNA, were significantly lower than the Negative Controls without siRNA. The Negative Control-I group that was challenged by Varroa mite and received no siRNA reached 100% IAPV infection at day 10 post experimental setup. In the Treatment-II group, which was not exposed to Varroa mites, the siRNA treatment resulted in a very low or even undetected viral infectivity at day 10 post treatment.

Figure 3:Results which indicate RNAi treatment could provide antiviral properties to bees when compared to control groups
Quantification of the IAPV titer in infected bees showed that bees in Treatment-II that received siRNA treatment and not exposed to Varroa mites had the lowest level of IAPV titer among four experimental groups. As a result, Treatment-II was chosen as a calibrator. The viral concentration of other groups was compared with calibrator and expressed as n-fold change. For bees from the Negative Control-I group challenged by Varroa mites and lacking siRNA treatment, the IAPV titer steadily increased over the whole experiment period and reached a maximum at day 10 post-treatment. The titer of IAPV in bees from Treatment-I was more than five-fold lower than in bees from Negative control-I (Figure 4). This result shows the feasibility of using siRNA to block or impair the translation of viral proteins to reduce virus replication, and reinforces the therapeutic potential of RNAi for treatment of honey bee diseases.
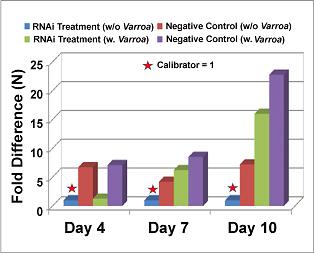
Figure 4: Results which show the viral concentration of bees over time, control groups had a 5 times greater concentration over RNAi treated groups
Summary
Since its discovery over a decade ago, remarkable progress has been made in unraveling the molecular mechanisms associated with RNAi. Despite great potential for this remarkable process in disease control, many challenges remain, ranging from potential side-effects for the bee hosts to delivery challenges and even counter-counter-attacks by the viral targets. These unsolved problems must be overcome before RNAi is used routinely as a therapeutic agent for diseases. As the pace of new findings and discoveries of applications continues to gain momentum and genomic information of honey bees and critical honey bee pests and pathogens accumulate, we anticipate that RNAi will be an effective and non-toxic therapeutic alternative for the treatment of bee diseases in the future.
References
Aronstein, K., Oppert, B., Lorenzen, M. D. 2011. RNAi in Agriculturally-Important Arthropods, in Book “RNA Processing” eds. by Grabowski, P. pp. 157-180. InTech Publishers.
Cox-Foster, D.L., Conlan, S., Holmes, E., Palacios, G., Evans, J.D., Moran, N.A., Quan, P.L., Briese, T., Hornig, M., Geiser, D.M., Martinson, V., van Engelsdorp, D., Kalkstein, A.L., Drysdale, A., Hui, J., Zhai, J., Cui, L., Hutchison, S.K., Simons, J. F., Egholm, M., Pettis, J. S., Lipkin W. I. 2007. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318: 283-287.
Evans, J.D., Chen, Y.P., Pettis, J., Williams, V. 2009. Bee cups: Single-use cages for honey bee experiments. J. Apicul. Res. 48: 300-302.
Fire, A., Xu, S. Q., Montgomery, M. K., Kostas, S. A., Driver, S. E., Mello, C. C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 (6669): 806–811.
Hunter, W., Ellis, J., vanEngelsdorp, D., Hayes, J., Westervelt, D., Glick, E., Williams, M., Sela, I., Maori, E., Pettis, J., Cox-Foster, D., Paldi, N. 2010. Large-scale field application of RNAi technology reducing Israeli acute paralysis virus disease in honey bees (Apis mellifera, Hymenoptera: Apidae). PLoS Pathogens 6 (12): e1001160.
Maori, E., Paldi, N., Shafir, S., Kalev, H., Tsur, E., Glick, E., Sela, I. 2009. IAPV, a bee-affecting virus associated with Colony Collapse Disorder can be silenced by dsRNA ingestion. Insect Molecular Biology . 18 (1): 55–60.
Paldi, N., Glick, E., Oliva, M., Zilberberg, Y., Aubin, L., Pettis, J. S., Chen, Y. P., Evans, J. D. 2010. Effective gene silencing of a microsporidian parasite associated with honey bee (Apis mellifera) colony declines. Appl Environ Microbiol 76: 5960–5964.
