A National Research and Extension Initiative to Reverse Pollinator Decline
This is part of an ongoing series of updates from the Managed Pollinator CAP. Additional installments can be found at the:
More information about the CAP can be found at:
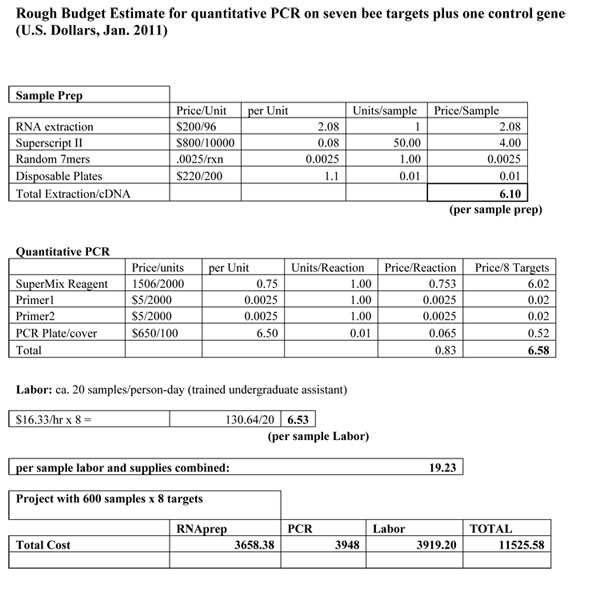
Protocol and budget for real-time PCR diagnostics for honey bees
Jay D. Evans USDA-ARS Bee Research Lab Beltsville, MD 20705 USA
Download a .pdf of this document
Sketched out below are the technical details for a routine we use regularly to generate genetic data from honey bee samples. These samples range from field collections of adult or larval bees to controlled experiments carried out on adult bees. The protocol focuses on the RNA of collected samples, a route that is essential for detecting the many bee viruses and one that also provides an opportunity to assess activity levels of specific honey bee or pathogen genes.
Acid Phenol RNA Extraction
Needed Reagents:
- Lysis buffer: 0.8M Guanidine thiocyanate, 0.4M Ammonium thiocyanate, 0.1M Sodium acetate, 5% glycerol, 2% Triton-X100. For 620 ml: 94.53g Guanidine thiocyanate,30.45g Ammonium thiocyanate, 33.4 ml 3M stock Sodium acetate, 50 ml glycerol, 20 ml Triton-X100, Read MSDS on thiocyanate safety. Mix in 300 ml water, bring up to 620 ml then autoclave. Use within 30 days.
- Acid Phenol (pH 4, Sigma Aldrich)
- Choloroform
- Isopropyl alcohol
- 70% ethanol
- Sterile, DNAse/RNAse free water
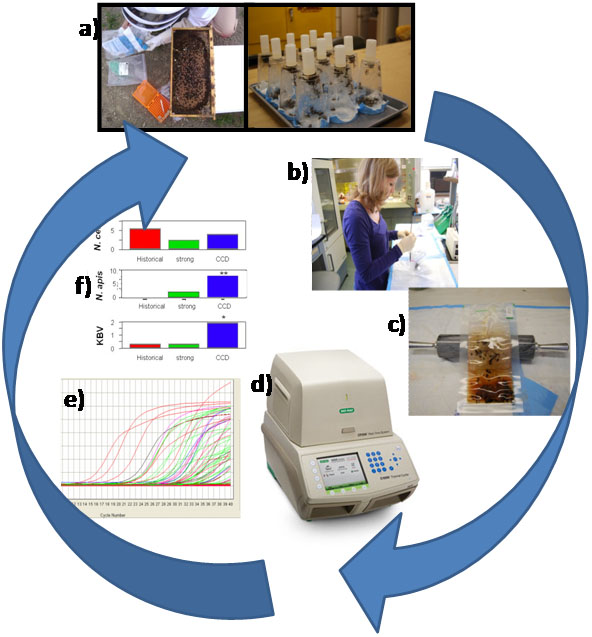
1. Collect ca. 200 bees from a field colony,or 30-50 from a cage experiment (Fig. 1A), into a 50 disposable centrifuge tube and flash freeze in liquid nitrogen, on dry ice, or in a -80°C freezer.
2. Count 50 bees from tube and place in a sealable bag (1 quart Zip-Loc or , BioReba, sample extraction bag; Fig. 1B)
3. Add 25 ml (or 500 μl/bee) Lysis buffer
4. Seal bag w/o air and smash with rolling pin until bees broken fully, for 30 sec (Fig. 1C)
5. Massage liquid to mix fully, then draw off 620 μl into a labeled microcentrifuge tube,avoiding bubbles
6. Freeze this aliquot immediately at -80°C
7. Thaw sample briefly, while still slightly frozen add 380 μl acid phenol, vortex 30 s,incubate at 95°C for ten min.
8. Cool to RT (can do on ice 5-10 min) and add 200 μl chloroform
9. Shake tubes vigorously by hand for 15 sec and then incubate at room temperature (RT) for 2 to 3 min.
10. Centrifuge the samples at 10,000 × g for 15 min at 4°C.
11. Collect upper aqueous phase into fresh tube containing 500 μl isopropyl alcohol, vortex 30 sec. (Can stop at this point, sample is stable for days at 4°C)
12. Incubate samples at 4°C for 30 min and centrifuge at 10,000 × g for 10 min at 4°C.
13. Remove the supernatant. Wash the RNA pellet once with 1 ml 70% ethanol
14. Mix the sample by vortexing and centrifuge at 7,000 × g for 5 min at 2 to 8°C.
15. Pour off supernatant, centrifuge 30 sec and pull off remaining supernatant with pipette
16. Briefly dry the RNA pellet (air-dry 5-10 min).
17. Dissolve RNA in 50 μl RNase-free water by incubating for 10 min at 55 to 60°C.
18. Store at -80°C
Complementary DNA (cDNA) synthesis
Needed Reagents:
- DNAse I (2 U/μl solution)
- RNAseOut (Invitrogen)
- DNTP (100mM solution)
- DNAse I Buffer (10X)
- Random primer set (7-mer at 10 mM concentration)
- Supercript II enzyme (200 U/μl; Invitrogen)
- Superscript Buffer (10X)
- DTT (0.1 M)
1. Prepare initial reaction mix containing, for each sample, 1 μl DNAse I solution, 0.5 μl primer mix, 0.4 μl RNAseOut, 0.4 μl dNTP mix, 1.1 μl DNAse buffer.
2. Aliquot 2.9 μl reaction mix per well
3. Add 8 μl of 1 μg/μl total RNA per well
4. Incubate 37°C 1 hour, then 75°C for 10 min, cool to 4°C
5. Heat plate to 70°C 10 min, place on ice
6. When cooled, centrifuge briefly
7. Incubate 42°C for 2 min
8. Add 4 μl Superscript mix to each sample (0.5 μl Superscript II enzyme, 1.5 μl Superscript buffer, 2 μl DTT)
9. Incubate 42°C 50 min, 70°C 15 min, 4°C
Quantitative PCR (20 μl reaction)
Needed Reagents:
- Sterile H2O
- Specific Forward and Reverse oligo primers
- Express SYBR Green ER qPCR SuperMix Universal (Buffer, Taq polymerase, dNTP’s).
It is of course common to mix qPCR reagents by hand, rather than buying a prepared mix, and numerous published papers describe such reaction mixes (e.g., one ‘home-made’ SYBR Green recipe that we have used for several years is described in Evans, 2006).)
1. Prepare Master Mix for each sample plus handling excess (calculate based on 100 samples for 96-well plate)
2. Aliquot into wells of plate, 19 μl each on ice
3. Add 1 μl template per well, on ice
4. Centrifuge plate to incorporate sample and remove bubbles
5. Program thermal cycler (see below), add plate, and run program (Fig. 1D)
6. Collect post-run data for spreadsheet manipulation and statistics (Fig. 1E, 1F).
- Programming thermal cycler: We use a Bio-Rad CFX-96 thermalcycler, programmed to start with a 50°C two minute step (this safety step enables the degradation of any possible past PCR products since the SuperMix incorporates dUTP into all PCR products and also includes Uracil DNA Glycosylase which will degrade any pre-existing uracil residues). Next is a 2 minute 95°C denaturing step, followed by 40 cycles of 95°C for 20 seconds, 60°C for 30 seconds, 72°C for 80 seconds. The fluorescence is measured at the 72°C step, since this provides another barrier against measuring short non-specific primer products (Evans, 2006).
Reference
Evans JD (2006) Beepath: An ordered quantitative-PCR array for exploring honey bee immunity and disease.